DICE Common Namespace
DICE Common Namespace <http://dice.nims.go.jp/ontology/> provides a URI (Uniform Resource Identifier) for machine-readable provision of DICE services.
Available Namespace
Aim of the DICE Common Namespace
The URI is meant to uniquely define terms used in each field. For example, for Materials Data Repository (MDR)'s work, "XAFS spectrum of Silver(I) sulfadiazine" (https://mdr.nims.go.jp/concern/datasets/5425kd364) , a unique URI (http://dice.nims.go.jp/ontology/mdr-ont#a57d4971-b66a-4a38-8aed-f0f357b2c328) could be assigned to a set of spectral data and metadata to allow the machine to recognize it. An important thing here is to connect this URI to resources outside of the MDR. There are many ways to do this, for example, one could assign a URI (http://dice.nims.go.jp/ontology/mdr-xafs-ont/Item#Q1298) to the measured sample (in this case, silver(I) sulfadiazine) and state that it is related to silver sulfadiazine in PubChem, a large and well-known database (https://pubchem.ncbi.nlm.nih.gov/). To do so, use the URI (http://rdf.ncbi.nlm.nih.gov/pubchem/compound/CID441244) in the namespace published by PubChem and write
wd:Q1298 skos:closeMatch compound:CID441244 .
Here, wd, skos, and compound are called prefix, each of which represents an abbreviated part of the URI, as shown below.
wd: <http://dice.nims.go.jp/ontology/mdr-xafs-ont/Item#>
skos: <http://www.w3.org/2004/02/skos/core#>
compound: <http://rdf.ncbi.nlm.nih.gov/pubchem/compound/>
With this sentence, even if work in MDR does not list chemical structures such as molecular weights or SMILES, we can integrate the information in PubChem. Thus, giving each resource a unique name (URI) and connecting that space to other resources will create greater knowledge, and that integration will bring forth new science.
It is an ontology that describes the concepts of words used in a resource, the relationships between words, and how words are used. To organize each expertise and define the connections to the outside world in each part. This is the aim of the DICE Common Namespace, which will be preceded by MDR and some databases, but will be extended as much as possible to other services aiming at open science.
The external ontologies and their prefixes used in DICE Common Namespace are as follows:
| Ontology | Name space | Prefix |
|---|---|---|
| DCMI | http://purl.org/dc/elements/1.1/ | dc |
| DCMI | http://purl.org/dc/terms/ | dcterms |
| OWL | http://www.w3.org/2002/07/owl# | owl |
| PRISM | http://prismstandard.org/namespaces/1.2/basic/ | prism |
| RDF | http://www.w3.org/1999/02/22-rdf-syntax-ns# | rdf |
| RDF Schema | http://www.w3.org/2000/01/rdf-schema# | rdfs |
| SKOS | http://www.w3.org/2004/02/skos/core# | skos |
| XML Schema | http://www.w3.org/2001/XMLSchema# | xsd |
mdr: <http://dice.nims.go.jp/ontology/mdr-ont#>
The data structure of mdr-ont is shown in the figure. Italics are concrete examples. The MDR Schema (https://github.com/nims-dpfc/mdr-schema) , which is published separately, shows the schema of metadata to be described in a "work", which is the unit of data set of MDR. The data structure shown here defines the positioning of the work within the overall MDR. Predicates are defined in the internationally accepted external ontology above, where the type of MDR work is a class (mdr:Works) and the URI of each work is linked as an instance of the class.
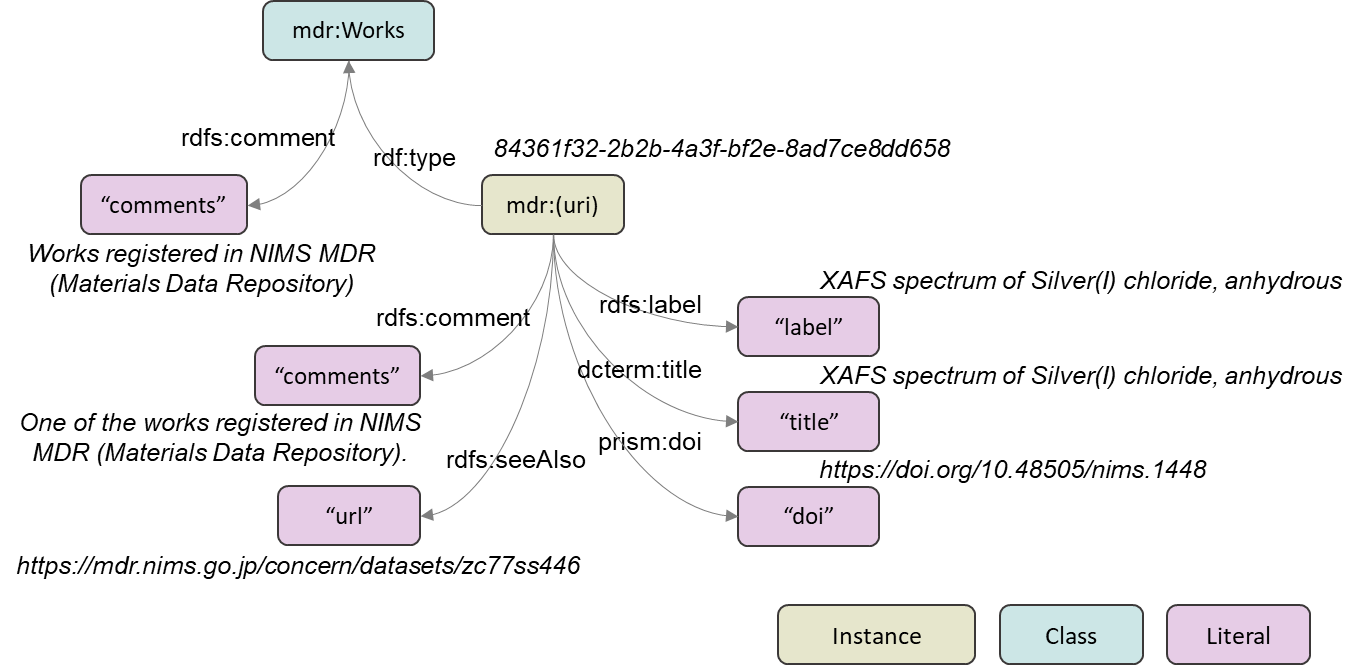
For semantic use of the data, including annotation details of the instances, please use the following RDF (Resource Description Framework) file in ttl format.
(Note: You may need to convert the extension to ".ttl" when uploading to a triple store.)
| File name (Click to download) | Version | Triples | Description |
|---|---|---|---|
| mdr-ont.owl | 0.3 | 15,495 | An ontology about MDR and its LOD. |
mdr-xafs-ont series: <http://dice.nims.go.jp/ontology/mdr-xafs-ont/>
The mdr-xafs-ont series defines a set of project namespaces that link the MDR XAFS DB (https://doi.org/10.48505/nims.1447) with external data and enable advanced searching.
In general, the data management required to implement a project involves (1) defining the vocabulary to be used, (2) defining the background physics and concepts, and (3) structuring the data in individual experiments (subprojects). These are defined in such a way that they do not contradict each other; the mdr-xafs-ont series creates a hierarchical namespace for them as follows.
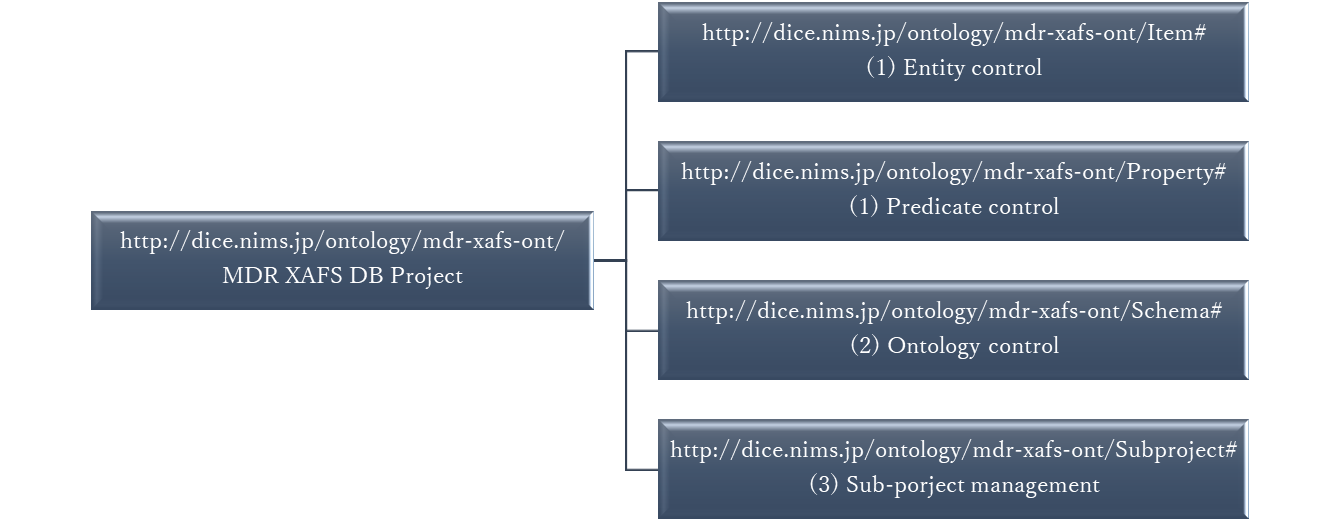
- http://dice.nims.go.jp/ontology/mdr-xafs-ont/Item#
- http://dice.nims.go.jp/ontology/mdr-xafs-ont/Property#
- http://dice.nims.go.jp/ontology/mdr-xafs-ont/Schema#
- http://dice.nims.go.jp/ontology/mdr-xafs-ont/Subproject#
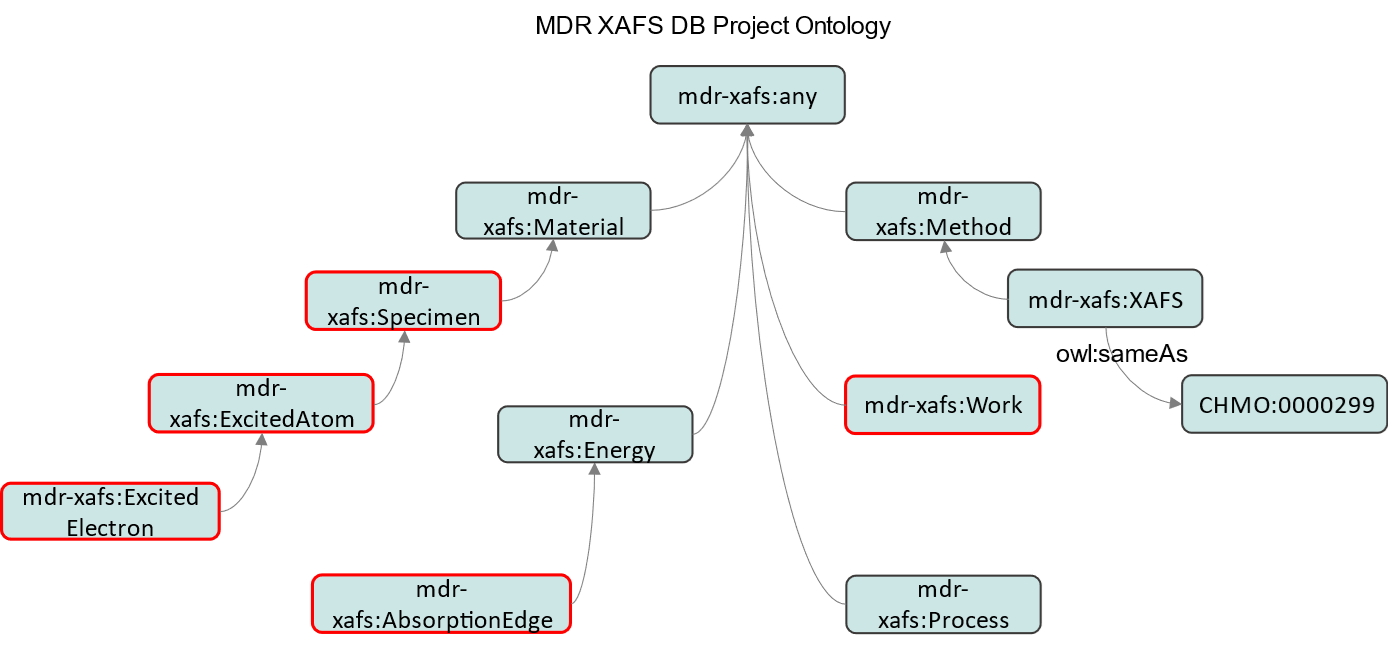
As shown in this figure, for convenience, the vocabulary in (1) is divided into entities and predicates. Vocabulary definitions will be managed equally as in a general dictionary, and thesaurus-like classifications and superordinate and subordinate concepts will be introduced in a different vocabulary management basis or in the schema (ontology) in (2). (2) is the structure of the physical concepts of the entities handled in this project, which is defined as shown in the following figure. Here, Chemical Methods Ontology, CHMO (http://purl.obolibrary.org/obo/) was referenced in the XAFS definition as an external ontology inherent to the mdr-xafs-ont series. This means that the XAFS handled in this project is linked to the external and can work with other measurements defined in CHMO. The red boxes indicate the classes used in the On Top of Single Figure (OTSF) subproject (The details will be discussed at https://mdr.nims.go.jp/concern/datasets/vh53wz94c), which is an example of (3) Subprojects.
Adding the predicates defined in (1) to this diagram, we obtain the following. That is, the predicates defined in (1) are used under the restrictions (rules for relating classes) described in (2).
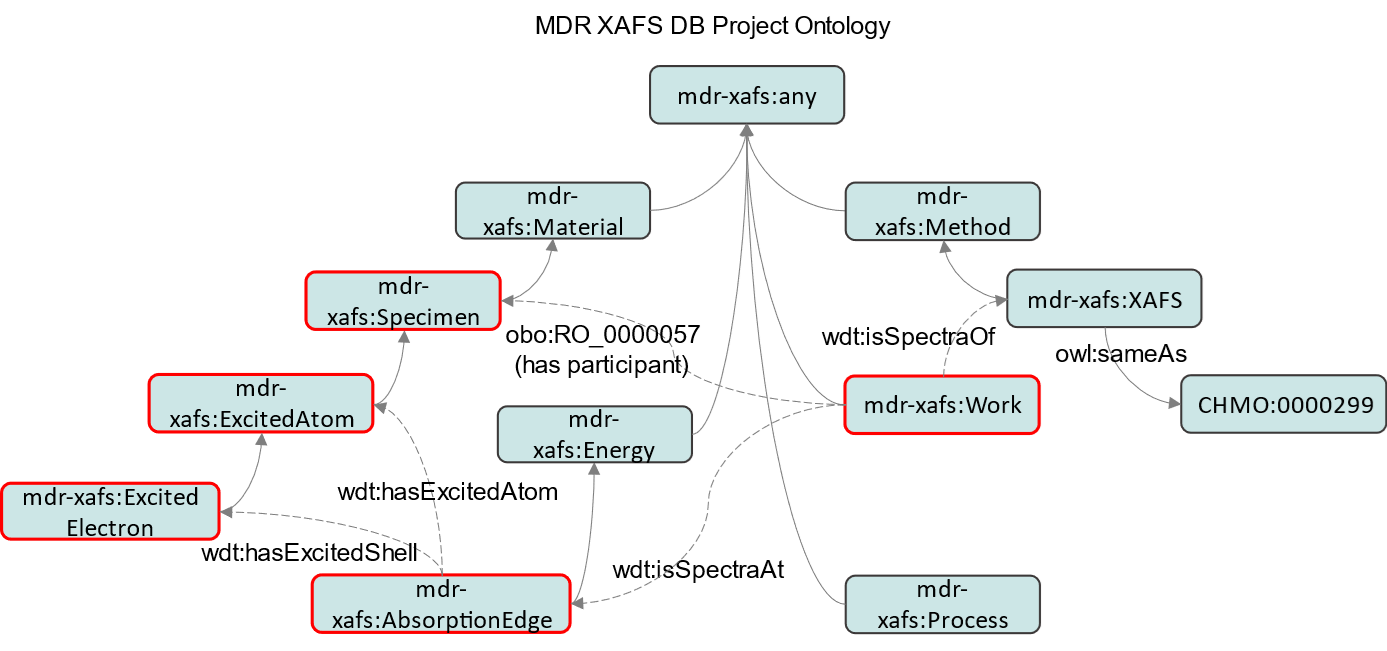
A machine-readable description of this figure can be found at http://dice.nims.go.jp/ontology/mdr-xafs-ont/Schema#.
(3) Data structuring in individual experiments (subprojects) is closed within the vocabulary and ontology defined in (1) and (2), so no new namespace is needed except for the definition of the subproject itself, http://dice.nims.go.jp/ontology/mdr-xafs-ont/Subproject#.
Each subproject is added to this space as an instance. If it is OTSF, it will be defined at
http://dice.nims.go.jp/ontology/mdr-xafs-ont/Subproject#OTSF
For semantic use of (1) through (3), please use the following RDF file in ttl format.
(Note: You may need to convert the extension to ".ttl" when uploading to a triple store.)
| File name (Click to download) | Version | Triples | Description |
|---|---|---|---|
| mdr-xafs-ont_Item.owl | 0.3 | 6,226 | Namespace for entities used in MDR XAFS DB (https://doi.org/10.48505/nims.1447). |
| mdr-xafs-ont_Property.owl | 0.2 | 29 | Namespace for predicates used in MDR XAFS DB (https://doi.org/10.48505/nims.1447). |
| mdr-xafs-ont_Schema.owl | 0.3 | 96 | An ontology about MDR XAFS DB and its LOD. |
| mdr-xafs-ont_Subproject.owl | 0.3 | 29 | Namespace for Sub-project of MDR XAFS DB (https://doi.org/10.48505/nims.1447). |
mdr-xps-ont series: <http://dice.nims.go.jp/ontology/mdr-xps-ont/>
The mdr-xps-ont series defines a set of namespaces for integrated use of the databases published by MDR, MDR HAXPES DB (https://doi.org/10.48505/nims.3056) and MDR XAFS DB (https://doi.org/10.48505/nims.1447).
For semantic use of the Schema, please use the following RDF file in ttl format.
(Note: You may need to convert the extension to ".ttl" when uploading to a triple store.)
| File name (Click to download) | Version | Triples | Description |
|---|---|---|---|
| mdr-xps-ont_Schema.owl | 0.1 | 96 | An ontology for MDR XAFS/HAXPES DB and their LOD. |
polymer-bio-ont series: <http://dice.nims.go.jp/ontology/polymer-bio-ont/>
The polymer-bio-ont series defines a set of project namespaces that allow PoLyInfo (https://polymer.nims.go.jp/) to be linked to external data and to allow advanced searching. In general, the data management required to implement a project involves (1) defining the vocabulary to be used, (2) defining the background physics and concepts, and (3) structuring the data in individual experiments (subprojects). These are defined in such a way that they do not contradict each other; the polymer-bio-ont series creates a hierarchical namespace for them as follows.
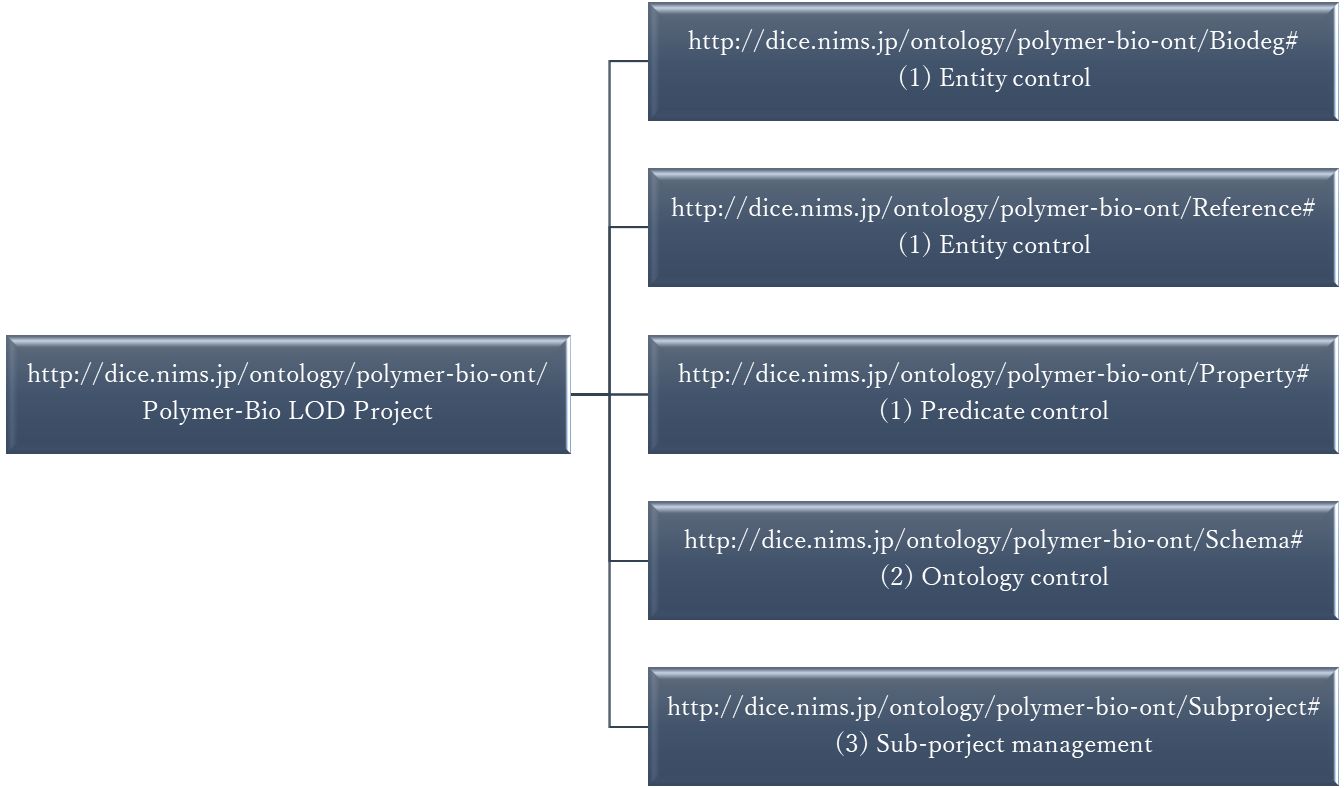
- http://dice.nims.jp/ontology/polymer-bio-ont/Biodeg#
- http://dice.nims.jp/ontology/polymer-bio-ont/Reference#
- http://dice.nims.jp/ontology/polymer-bio-ont/Property#
- http://dice.nims.go.jp/ontology/polymer-bio-ont/Schema#
- http://dice.nims.go.jp/ontology/polymer-bio-ont/Subproject#
As shown in this figure, for convenience, the vocabulary in (1) is divided into entities and predicates. Entities in this project are specialized to biodegradation of synthetic polymers, so they are grouped in Biodeg# as the vocabulary of the domain. The bibliography of the information source is provided in Reference#, machine-readable format. (2) is the structure of the physical concept of biodegradation covered in this project, as shown in the figure below. The distinctive feature of this concept is that all complex parts, such as the definition of biodegradation, its position as a process, the bacteria species involved, polymers, and so on, are left to an external ontology. One of the goals of this project is to establish a linkage between these large concepts and to establish collaboration among the different fields. The definition of biodegradation of synthetic polymers is based on the Environment Ontology, ENVO (http://purl.obolibrary.org/obo/).
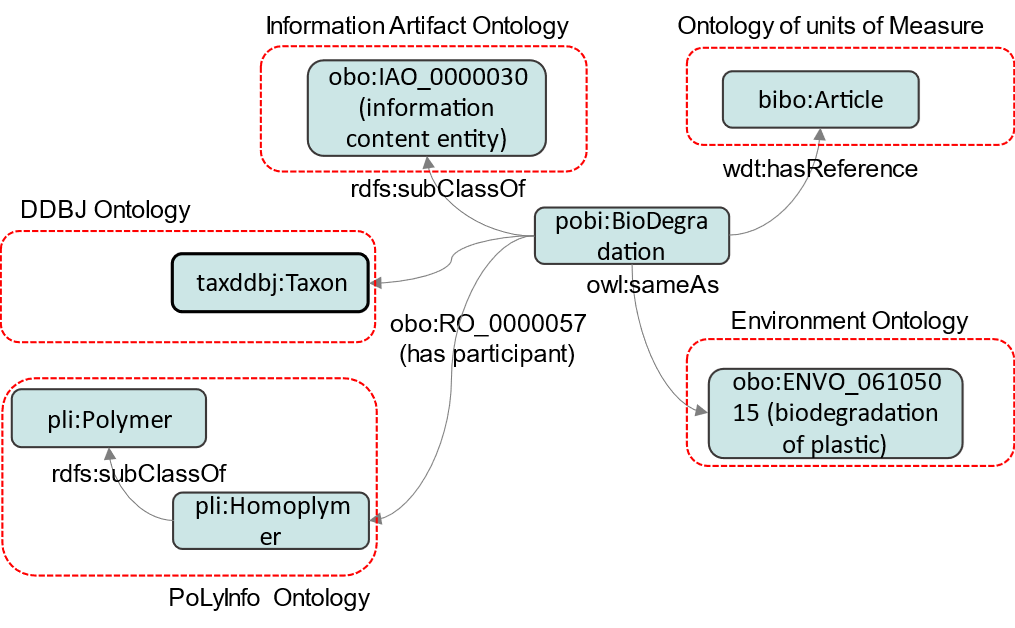
The machine-readable description of this diagram can be found at http://dice.nims.go.jp/ontology/polymer-bio-ont/Schema#.
The external ontology used specific to this project is as follows
| Ontology | Name space | Prefix |
|---|---|---|
| Bibo | http://purl.org/ontology/bibo/ | bibo |
| Obo | http://purl.obolibrary.org/obo/ | obo |
| PoLyInfo | http://dice.nims.go.jp/ontology/PoLyInfo-ont/Schema# | pli |
| Taxddbj | http://www.w3.org/2002/07/owl# | taxddbj |
| Taxonomy | http://identifiers.org/taxonomy/ | taxonomy |
Data structuring in individual experiments (subprojects) is restricted to the vocabulary and ontology defined in (1) and (2), so no new namespace is required except for the definition of subprojects, http://dice.nims.go.jp/ontology/polymer-bio-ont/Subproject#.
Each subproject is added to this namespace as an instance. A subproject BDG-S, which represents biodegradation using bacterial species, would be denoted by
http://dice.nims.go.jp/ontology/polymer-bio-ont/Subproject#BDG-S.
For semantic use of (1) through (3), please use the following RDF file in ttl format.
(Note: You may need to convert the extension to ".ttl" when uploading to a triple store.)
RDF based on the polymer-bio-ont series will be published at https://integbio.jp/rdf/.
| File name (Click to download) | Version | Triples | Description |
|---|---|---|---|
| polymer-bio-ont_Biodeg.owl | 0.1 | 1506 | Definition of bio-degradation of polymers registered in PoLyInfo. |
| polymer-bio-ont_Reference.owl | 0.1 | 1035 | References for bio-degradation of polymers registered in PoLyInfo. |
| polymer-bio-ont_Property.owl | 0.1 | 15 | Namespace for predicates used in polymer-bio-ont. |
| polymer-bio-ont_Schema.owl | 0.1 | 49 | An ontology for polymer-bio linked data. |
| polymer-bio-ont_Subproject.owl | 0.1 | 18 | Namespace for Sub-project of polymer-bio linked data. |
PoLyInfo-ont series: <http://dice.nims.go.jp/ontology/PoLyInfo-ont/>
The PoLyInfo-ont series provides a namespace for describing PoLyInfo (https://polymer.nims.go.jp/) as a polymer concept.
Details are available on the MDR at https://doi.org/10.48505/nims.4413.
Contact
For inquiries about DICE Common Namespace, please use DICE Contact Form.
(Please select "Not specified" for the service item.)
